Abstract
Free full text

Characterization and Comparison of Recombinant Simian Immunodeficiency Virus from Drill (Mandrillus leucophaeus) and Mandrill (Mandrillus sphinx) Isolates
Abstract
Since simian immunodeficiency virus (SIV) was found to be the source of the human AIDS pandemic, a major goal has been to characterize the diversity of SIV strains in the wild and to assess their potential for crossover into humans. In the present study, SIV was isolated from a seropositive drill (Mandrillus leucophaeus) and three seropositive mandrills (Mandrillus sphinx) by using macaque peripheral blood mononuclear cells (PBMC). Full-length sequences were obtained from a drill and mandrill and designated SIVdrl1FAO and SIVmnd5440, respectively. A 182-bp fragment of the pol genes of the two remaining mandrill SIV isolates was also analyzed. Phylogenetic analyses demonstrated that SIVdrl1FAO formed a monophyletic clade with SIVmnd5440 and SIVmndM14, recently designated SIVmnd type 2. Both the SIVdrl and SIVmnd type 2 genomes carried a vpx gene and appeared to share a common ancestor with SIVrcm in the 5′ region of the genome and with SIVmndGB1 (type 1) in the 3′ region of the genome. A statistically significant recombination breakpoint was detected at the beginning of envelope, suggesting that the viruses were descendents of the same recombinant. Phylogenetic analysis of vpx and vpr genes demonstrated that the vpx genes formed a monophyletic cluster that grouped with vpr from SIVagm. In addition, both SIVdrl1FAO and SIVmnd5440 replicated in human PBMC and therefore could pose a risk of transmission to the human population.
Since human immunodeficiency virus type 1 (HIV-1) and HIV-2 have been shown to originate from simian immunodeficiency virus (SIV) in African primates (9, 17, 40, 41), characterizing these viruses has become a major goal of AIDS research. To date, more than 20 species of SIV-infected African nonhuman primates have been detected (4, 18, 31, 37, 39); 34 complete sequences of these viruses have been published (30). SIV and HIV are classified as primate lentiviruses (PLVs), and at the present time six lineages, based upon phylogenetic relationships of full-length genomes, are recognized (5, 12). These six designated lineages are (i) SIVcpz from chimpanzees (Pan troglodytes), including HIV-1 (17, 25, 26, 41, 55); (ii) SIVsm from sooty mangabeys (Cercocebus atys), including HIV-2 and SIVmac from captive macaques (Macaca spp.) (21, 24, 36); (iii) SIVagm from African green monkeys (members of the Chlorocebus aethiops superspecies) (1, 2, 14-16, 23, 27, 29, 35); (iv) SIVsyk from Sykes' monkeys (Cercopithecus albogularis); (v) SIVlhoest from L'Hoest monkeys (Cercopithecus lhoesti), including SIVsun from sun-tailed monkeys (Cercopithecus solatus) and SIVmnd type 1 (GB1) from mandrills (Mandrillus sphinx) (3, 4, 22, 54); and (vi) SIVcol from Colobus monkeys (Colobus guereza) (12). In addition, another novel recombinant SIV from greater spot-nosed monkeys (Cercopithecus nicitans) was recently described (13). SIVgsn shares the vpu gene of HIV-1 and SIVcpz and is most closely related to these viruses in the envelope but clusters with SIVsyk in Gag and part of Pol. Some SIV strains, such as SIVdrl from drills (Mandrillus leucophaeus), SIVtal from talapoin monkeys (Miopithecus talapoin), SIVmus from mustached guenons (Cercopithecus cephus), SIVmon from Mona monkeys (Cercopithecus mona), and SIVdeb from DeBrazza monkeys (Cercopithecus neglectus) have only been partially characterized (11, 38, 39). Sequencing of the entire genome is necessary to classify those viruses into a particular lineage or to designate them as members of a novel lineage.
Phylogenetic studies of nonhuman PLVs have suggested that the divergence of viral lineages reflected in some cases the divergence of their host lineages, suggesting host-dependent evolution (1, 4, 44, 45). However, in many cases the differences between phylogenetic relationships of some viruses and primate phylogeny indicated that there have been multiple cross-species transmissions in the past (7, 28, 51). Cross-species transmission of PLVs can have a variety of outcomes in the new host. For example, if the recipient of the transmitted virus is already infected by a PLV, superinfection by other PLVs has the potential for generating recombinant viruses that could have a complex mosaic genome. Thus, some SIV isolates could not be classified into a particular lentivirus lineage because their genome was the result of one or more recombination events between viruses from different lineages. For example, SIVagm from sabaeus monkeys was the result of a recombination event between a SIVagm ancestor and another virus potentially from the SIVsm lineage (27). SIVrcm has been shown to cluster with different lentivirus lineages in different regions of the genome, but it is still uncertain whether SIVrcm or one of the potential parental viruses is the recombinant (6, 18). The most recently discovered mosaic virus was SIVmndM14, which was found to be recombinant between SIVrcm and SIVmndGB1 (48).
Based on the commonly accepted hypothesis that HIV-1 and HIV-2 were the result of multiple cross-species transmissions to humans from chimpanzees and sooty mangabeys, respectively (9, 10, 17, 40, 41), it is important to consider the possibility of ongoing zoonotic transfer. People in west central Africa are still routinely exposed to a large variety of genetically divergent SIV stains (20, 39). Indeed, epidemiological surveys identified a human serum sample from Cameroon that showed a high reactivity directed solely and strongly against the SIVmnd V3 loop, raising the possibility that mandrills may represent a viral reservoir for humans similar to sooty mangabeys in western Africa and chimpanzees in central Africa (48). Moreover, a variety of SIV strains are able to replicate in human peripheral blood mononuclear cells (PBMC) and macrophages in vitro, indicating the possibility for another crossover of SIV into the human population (6, 19).
In the present study, we isolated SIV from blood samples from one drill and three mandrills housed in North American and European zoos. The highly endangered drill is a large, short-tailed monkey restricted in the wild to several blocks of forest between the Cross river in southeastern Nigeria, the Sanaga river in southwestern Cameroon, and the island of Bioko (Equatorial Guinea) (11). The related mandrill is also a large semiterrestrial primate that lives in the tropical rain forests of Cameroon and Gabon (48). Our goal was to isolate and molecularly characterize SIV from a drill and mandrill and evaluate their in vitro host ranges and coreceptor usages. For this purpose, we characterized the full-length genomes of the SIV isolates from the drill (no. 1FAO) and from one of the mandrills (no. 5440). We also determined the host range of SIVdrl1FAO and SIVmnd5440 in permanent cell lines and in primary PBMC and evaluated the coreceptor usage of SIVdrl1FAO.
MATERIALS AND METHODS
Primate samples.
Fresh blood specimens were obtained from a drill (1FA0) and three mandrills (5440, Pearl, and Gunnite) housed at North American or European zoological parks in accordance with the guidelines of the animal care and use committees at their institutions. The single male drill was in an exhibit that was exposed to a wild-born drill with an unknown SIV serostatus. Mandrill 5440 [female] was captive born to African-born parents with unknown SIV serostatus. The countries of origin of the wild-born animals are not known. Mandrill Gunnite [male] and Pearl [female] were born in captivity to captive-born parents of which the mother was SIV seropositive in both cases. From the four animals, PBMC and plasma were obtained by standard Ficoll-Hypaque density gradient centrifugation and stored in liquid nitrogen until use.
Virus isolation.
After being thawed, drill and mandrill PBMC were washed twice with Hanks' buffered salt solution and stimulated with 2 μg of phytohemagglutinin (PHA) (Sigma, St. Louis, Mo.) per ml for 3 days. PBMC from SIV-negative pig-tailed macaques were separated by Ficoll-Hypaque density gradient centrifugation of whole blood and also stimulated with 2 μg of PHA per ml for 3 days. PBMC blasts from the SIV-positive drill and three SIV-positive mandrills (5440, Pearl, and Gunnite) were cocultivated with 2 × 107 PBMC blasts from pig-tailed macaques and maintained in complete RPMI (RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum, 2 mM glutamine, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 10 mM HEPES) supplemented with 5 half-maximal units of human interleukin-2 (Advanced Biotechnologies, Columbia, Md.) per ml. Supernatants were sampled every three days and subjected to reverse transcriptase (RT) assay. Supernatants were harvested at the peak of RT activity, filtered through a 0.45-μm-pore-size filter, and cryopreserved as virus stock. Virus stocks were designated SIVdrl1FAO, SIVmnd5440, SIVmndPea, and SIVmndGun.
PCR amplification of the viral genome.
PCRs were performed with DNA extracted from PBMC by using phenol-chloroform or from cell lysates as described previously (49). To generate a small fragment of 194 bp in the RT portion of the pol gene from cell lysate of drill, degenerate primers DR1, DR2, DR4, and DR5 were used as previously described (11). Amplification was performed with an ExTaq kit (Takara, Otsu, Shiga, Japan) with an initial denaturation at 94°C for 2 min, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a final extension of 72°C for 10 min. Primer DR1 was used at 0.8 pmol per μl, and DR2, DR4, and DR5 were used at 2.5 pmol per μl. A second fragment of 182 bp located in the integrase part of the pol gene was generated by nested PCR with primers mpolF1, mpolR1, mpolF2, and mpolR2 (Table (Table1).1). The conditions for PCR amplification were the same as described above.
TABLE 1.
Nested primer pairs and their genomic locations
| Primer | Sequence (5′ to 3′) | Specificity | Amplified fragment (bp) | Gene | Location (nt) |
|---|---|---|---|---|---|
| mpolF1 | ATTATAGAACAGGTTAGAGATCAGGCA | SIV | 233 | pol (outer set) | 4272-4297a |
| mpolR1 | GGTCCTTTCCACTGTTGATCCCTT | 4480-4503a | |||
| mpolF2 | TTAGAGACAGCAGTTCAAATGGCAGTA | 182 | pol (inner set) | 4304-4330a | |
| mpolR2 | TCCCTTCCTTCTCTGTAATAAACCCGA | 4459-4485a | |||
| drlF1 | TGGCGCCCGAACAGGGACTTGA | SIVdrl1FAO | 4480 | PBS | 1-22 |
| mpolR1 | GGTCCTTTCCACTGTTGATTCCTT | pol | 4480-4503 | ||
| drlF2 | GCATACCACAGGAGTTCCCTATAA | 5935 | pol | 4204-4227 | |
| drlR2 | GTTCTTTCGCTACCCAGACAAGAT | gag | 388-406 | ||
| mnd5440F1 | TGGCGCCCGAACAGGGACTTGA | SIVmnd5440 | 4485 | PBS | 1-22 |
| mnd5440R1 | TCCCTTCCTTCTCTGTAATAAACCCGA | pol | 4459-4485 | ||
| mnd5440F2 | GGCTAGAAACAGCAGTGCAAATGG | 5457 | pol | 4302-4325 | |
| mnd5440R2 | GTCCCTTACAGGTGGGTGCGACCTGAA | PBS | 29-55 |
To amplify the entire SIVdrl1FAO or SIVmnd5440 genomes, genomic DNA was extracted from infected Molt4clone8 cells by using phenol-chloroform. Two overlapping fragments, from the primer binding site (PBS) to integrase (4.5 kb) and from integrase to the PBS-gag gene (5.5 and 5.9 kb, respectively), were amplified by targeting the integrated genomic as well as unintegrated circular DNA. The primer sequences and location of the expected amplified product are described in Table Table1.1. PCRs were performed in a volume of 50 μl with the Expand Long Template High Fidelity PCR system (Roche Molecular Biochemicals) under the following conditions: a hot start at 94°C for 3 min; 10 cycles of denaturation at 94°C for 20 s, annealing at 58°C for 30 s, and extension at 68°C for 5 to 7 min; and 20 cycles with extension at 68°C for 5 to 7 min with an increment of 20 s per cycle. Amplification was completed by a final extension at 72°C for 15 min. The entire 50-μl PCR product was separated on a 0.9% agarose gel. Bands of the correct size were excised, cloned into pCRII-TOPO vector (TOPO TA Cloning kit; Invitrogen, Carlsbad, Calif.) and sequenced by automated fluorescent sequencing (Taq amplification and termination; Perkin-Elmer Applied Biosystems, Warrington, United Kingdom).
Sequence analysis.
The sequences of SIVdrl1FAO and SIVmnd5440 were aligned and compared to representatives of the major lentivirus lineages, including some of those in the HIV sequence database at http://hiv-web.lanl.gov/ALIGN_CURRENT/ALIGN-INDEX.html (“other SIV complete genome DNA” alignment). For some of the phylogenetic analyses, third-codon positions were removed. Columns in the alignment, in which gaps had been inserted to maintain the alignment through regions with insertions and deletions, were stripped prior to the analyses. Protein identities were calculated from the gap-stripped alignment by using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The gap-stripped alignment was further analyzed with the SIMPLOT program from Stuart Ray (http://www.med.jhu.edu/deptmed/sray/download), both as nucleotide and as translated amino acid sequences, in order to determine the regions of the alignment to be analyzed separately by phylogenetic methods. A neighbor-joining phylogenetic tree was built for each genomic region, using the PHYLIP, DNADIST, and NEIGHBOR programs (http://evolution.genetics.washington.edu/phylip.html) with the F84 model of evolution. The resulting tree was used as input, along with the alignment, to Gary Olsen's DNArates program (http://geta.life.uiuc.edu/~gary/programs/DNArates.html) to compute site-specific rates of evolution. These rates were then included in the input to a modified version of fastDNAml written by Tanmoy Bhattacharya (http://www.santafe.edu/~btk/science-paper/bette.html), which computes maximum-likelihood trees incorporating site-specific rates of evolution. Bootstrap support (100 replicates) for each of the trees was calculated by using the PHYLIP SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE programs. Although the bootstrap trees were computed via the neighbor-joining method, rather than by maximum likelihood with site-specific rates, the topologies were often identical although branch lengths differed significantly. Trees were edited in TreeTool (ftp://ftp.cme.msu.edu/pub/RDP/programs/TreeTool/).
Infection of chimpanzee PBMC, human PBMC, and human CD4+-T-cell lines with SIVdrl1FAO or SIVmnd5440.
To test the in vitro host range of SIVdrl1FAO and SIVmnd5440, human and chimpanzee PBMC and a panel of CD4+ human cells were used. PBMC were separated from heparinized whole blood of HIV-negative humans and SIV- and HIV-negative chimpanzees by Ficoll-Hypaque density gradient centrifugation. PBMC were stimulated with 2 μg of PHA (Sigma) per ml in complete RPMI 1640 with 10% fetal calf serum, supplemented with 5 half-maximal units of human interleukin-2 per ml, for 3 days before viral inoculation. The human CD4+ cell lines CEMx174, CEM-SS, C8166, H9, Hut78, MT-2, MT-4, Molt4clone8, PM1, SupT1, and U937 were maintained in RPMI complete medium. Five million PHA-stimulated PBMC or 2 million human CD4+ T cells were pelleted, and the cell pellet was incubated with 1 ml of SIVdrl1FAO or SIVmnd5440 (approximately 500,000 cpm of RT activity) for 2 h with agitation every 30 min. The cells were washed once with medium, 5 ml of complete RPMI was added, and the cells were transferred to T25 flasks (Nalge Nunc International, Rochester, N.Y.). Cell culture supernatants were collected every 3 days, and the infection of the cells was monitored for up to 6 weeks.
GHOST cell assay.
The coreceptor usages of SIVdrl1FAO and SIVmnd5440 were determined by using the GHOST cell assay (34). The GHOST cell panel was obtained through the AIDS Reagent and Reference Program, National Institutes of Health (Rockville, Md.). Cells (2 × 104) from each of the cell lines were seeded in 24-well plates (Costar, Coring, N.Y.) and cultivated for 2 days prior to infection. One milliliter of virus stock (corresponding to approximately 500,000 cpm of RT activity) was gently added to the cells, and after 4 h of incubation at 37°C, the cells were washed once with medium, followed by the addition of 1 ml of fresh medium. Every other day the cells were split 1:2 and 0.5 ml of culture supernatant was collected. The production of SIV Gag p27 antigen was measured by using the RETRO-TEK SIV-1 p27 antigen enzyme-linked immunosorbent assay (ZeptoMetrix Corp., Buffalo, N.Y.) according to the manufacturer's instructions.
Nucleotide sequence accession numbers.
The complete genomes of SIVdrl1FAO and SIVmnd5440 have been submitted to GenBank under accession numbers AY159321 and AY159322.
RESULTS
Identification and isolation of SIVdrl and SIVmnd.
Antibodies to SIV were detected using a commercially available HIV-2 Western blot kit (HIV-2 blot1.2; Genelabs, Singapore). Sera of mandrill 5440 and drill FAO cross-reacted with Env gp36, Env gp80, and Gag p26 proteins of HIV-2 (Fig. (Fig.1).1). Virus was isolated from PBMC of the seropositive drill and from all three seropositive mandrills (Gunnite, Pearl, and 5440) by cocultivation with PHA-stimulated pig-tailed macaque PBMC. Virus was isolated from the SIV-positive mandrills after 5 weeks of coculture, with peak RT activity by 6 weeks postinfection. Cytopathic effects were not observed during the entire period of coculture. A similar strategy was used for isolation of SIVdrl1FAO, except that isolation only required 1 week of coculture.
Genomic organizations of SIVdrl and SIVmnd.
Partial pol sequences were amplified from three SIV-positive mandrills and one SIV-positive drill by using degenerate primers as detailed in Materials and Methods. Subsequently, the complete genomes of SIVdrl1FAO and SIVmnd5440 were amplified in two fragments (4.5 kb and 5.5 to 5.9 kb) by targeting the integrated genomic as well as unintegrated circular DNA. The PCR fragments were cloned by using the TOPO cloning system (Invitrogen) and sequenced, and the sequences assembled to obtain the full-length genomes, which were compared to those of other PLVs. The proviral genomes of SIVdrl1FAO and SIVmnd5440 shared their genomic organization with SIVmndM14, SIVrcm, SIVsm, SIVmac, and HIV-2. These viruses possess a vpx gene in addition to vpr but lack a vpu gene which is present in the SIVcpz/HIV-1 lineage and in SIV from greater spot-nosed and mona monkeys (13; J. Clewley, 9th International Workshop on HIV Dynamics and Evolution, abstr. 5, 2002).
SIVdrl1FAO and SIVmnd5440 share the same mosaic structure.
We compared protein identities of SIVdrl1FAO and SIVmnd5440 to published full-length sequences of SIV in order to estimate their relationship to other PLVs. SIVdrl1FAO was most closely related to SIVdrl/Clewley (unpublished; GenBank accession no. AJ310481), with identities of 92 and 93% in partial fragments of Gag and Pol, respectively (Table (Table2).2). Next, SIVdrl1FAO was most closely related throughout the entire genome to SIVmnd5440 and SIVmndM14, with a protein identity of 81 to 83% in Gag and Pol. In comparison to the other PLVs, SIVdrl1FAO was most closely related in the Gag, Pol, Vif, Vpx, Vpr, and Tat proteins to SIVrcm in Env and Nef to SIVmnd type 1 (SIVmndGB1) and in Rev to SIVlhoest. SIVdrl1FAO was also closely related to SIVagmSAB in Gag and Pol, probably due to the relationship between SIVrcm and SIVagmSAB. SIVdrl1FAO was only distantly related to viruses of the other primate lentivirus lineages.
TABLE 2.
Protein sequence identities between SIVdrl1FAO and other PLVs
| Virus (accession no.) | % Identity with SIVdrl1FAO in:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gag | Pol | Vif | Vpx | Vpr | Tata | Revb | Env | Nef | |
| SIVdrl/Clewley (AJ310481) | 92c | 93d | |||||||
| SIVmnd5440 (AY159322) | 83 | 81 | 69 | 70 | 78 | 52 | 57 | 67 | 66 |
| SIVmndM14 (AF328295) | 81 | 81 | 70 | 68 | 80 | 52 | 61 | 72 | 63 |
| SIVrcm (AF349680) | 73 | 77 | 51 | 57 | 63 | 46 | 39 | 38 | 41 |
| SIVcpz (X52154) | 59 | 67 | 42 | 49 | 37 | 28 | 36 | 40 | |
| SIVsm (L09212) | 64 | 61 | 44 | 42 | 63 | 39 | 48 | 40 | 43 |
| SIVagmSab (U04005) | 72 | 72 | 49 | 42 | 39 | 39 | 37 | 48 | |
| SIVagmVer (L40990) | 67 | 66 | 46 | 42 | 44 | 43 | 38 | 44e | |
| SIVlhoest (AF075269) | 50 | 58 | 35 | 44 | 37 | 50 | 55 | 45 | |
| SIVsyk (L06042) | 60 | 55 | 41 | 25 | 50 | 30 | 38 | 39 | |
| SIVcol (AF301156) | 47 | 51 | 22 | 28 | 36 | 30 | 29 | 31 | |
| SIVmndGB1 (M27470) | 54 | 58 | 36 | 40 | 44 | 37 | 63 | 55 | |
SIVmnd5440 showed a pattern of genetic relationships similar to that of SIVdrl1FAO (Table (Table3).3). It was closely related to SIVmndM14, with >90% identity in Gag and Pol and an overall protein identity of at least 74%, indicating that both SIV isolates represented the same type of virus. Next, SIVmnd5440 was most closely related to SIVdrl1FAO and also to SIVrcm in the 5′part of the genome and to SIVmndGB1 in the 3′ part of the genome. The relationship of SIVdrl1FAO and SIVmnd5440 to SIVrcm from the Gag to the Tat protein and to SIVmndGB1 in Env and Nef and the overall identity to SIVmndM14 across the entire genome indicated that both novel SIV isolates might have been generated by recombination.
TABLE 3.
Protein sequence identities between SIVmnd5440 and other PLVs
| Virus (accession no.) | % Identity with SIVmnd5440 in:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gag | Pol | Vif | Vpx | Vpr | Tata | Revb | Env | Nef | |
| SIVmndM14 (AF328295) | 93 | 91 | 75 | 83 | 85 | 74 | 76 | 80 | 78 |
| SIVdrl1FAO (AY159321) | 83 | 81 | 69 | 70 | 78 | 52 | 57 | 67 | 66 |
| SIVrcm (AF349680) | 70 | 77 | 49 | 63 | 66 | 54 | 30 | 36 | 46 |
| SIVcpz (X52154) | 56 | 67 | 40 | 52 | 37 | 30 | 35 | 40 | |
| SIVsm (L09212) | 65 | 59 | 40 | 39 | 64 | 43 | 39 | 38 | 48 |
| SIVagmSab (U04005) | 71 | 70 | 47 | 42 | 41 | 43 | 36 | 48 | |
| SIVagmVer (L40990) | 65 | 65 | 47 | 40 | 54 | 41 | 38 | 43c | |
| SIVlhoest (AF075269) | 51 | 58 | 34 | 44 | 44 | 50 | 49 | 45 | |
| SIVsyk (L06042) | 60 | 54 | 40 | 25 | 48 | 30 | 35 | 38 | |
| SIVcol (AF301156) | 47 | 50 | 25 | 27 | 33 | 28 | 30 | 30 | |
| SIVmndGB1 (M27470) | 53 | 61 | 36 | 44 | 39 | 46 | 58 | 54 | |
In order to more thoroughly investigate the relationship of SIVdrl1FAO and SIVmnd5440 to other PLVs and to rule out evidence of recombination, similarity plots were constructed. As shown in Fig. Fig.2A,2A, SIVdrl1FAO was compared to partial gag-pol sequences of SIVdrl/Clewley, SIVmnd5440, SIVmndM14, SIVmndGB1, and SIVrcm. As expected from the amino acid identities, SIVdrl1FAO was most closely related to SIVdrl/Clewley in the gag-pol region. The relationship between the two SIVdrl strains appeared to be similar to the relationship between SIVmnd5440 and SIVmndM14.
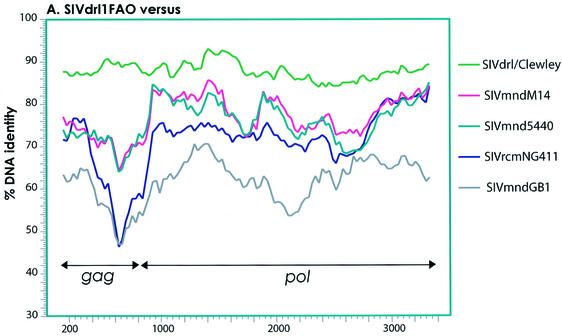

Similarity plots comparing a 3,450-bp gap-stripped partial gag-pol alignment of SIVdrl1FAO with other SIV isolates from drill and mandrill (A), comparing the full-length genome of SIVdrl1FAO with SIVmnd5440, SIVmndM14, SIVrcm, and representatives of the other major PLV lineages (B), and comparing the full-length genome of SIVmnd5440 with SIVdrl1FAO, SIVmndM14, SIVrcm, and representatives of the other major PLV lineages (C). The gene boundaries are indicated by arrows. In the Simplots for panels B and C, third-codon positions of the alignment have been removed. The fractional nucleotide sequence difference was calculated for a window size of 300 bp (A) or 400 bp (B and C) moved in steps of 30 bp (A) or 40 bp (B and C) with Hamming distances.
We also constructed similarity plots to compare SIVdrl1FAO and SIVmnd5440 to full-length genomes of SIV. As shown in Fig. Fig.2B,2B, SIVdrl1FAO was most closely related to SIVmnd5440 and SIVmndM14 across the entire genome, indicating a common origin of both viruses. As already indicated by the protein identities, SIVdrl1FAO and SIVmnd5440 were most closely related to SIVrcm in gag, pol, vif, vpx, and vpr, whereas in env they were most closely related to SIVmndGB1 (Fig. 2B and C). This observation confirmed that the genomes of SIVdrl1FAO and SIVmnd5440 were recombinant.
A phylogenetic tree of a small pol fragment (Fig. (Fig.3A)3A) demonstrated that SIVmnd type 2 and SIVdrl sequences formed monophyletic clusters, with the exception of SIVmnd2-99CM32 which fell in the SIVdrl cluster. Since the host of SIVmnd2-99CM32 was a wild-born pet monkey genetically confirmed to be a mandrill by mitochondrial sequencing (50), some cross-species transmission of SIVdrl to a mandrill must have been occurred in the past. This is particularly interesting, because the habitats of drills and mandrills are located north and south of the Sanaga River, respectively, and do not overlap. Therefore, transmission of SIV between drill and mandrill is an unexpected finding.


Phylogenetic trees of PLV full-length genomes. (A) Unrooted phylogenetic tree (maximum likelihood with site-specific substitution rates) of a conserved fragment of the pol gene, including many of the available SIVmnd and SIVdrl sequences. The alignment was 397 nucleotides after gap stripping. SIVdrl1FAO and SIVmnd5440 are boxed. (B and C) The primate lentiviral genomes were aligned as described in Materials and Methods, third-codon positions were removed, and columns containing gaps were stripped from the alignment. (B) Representative tree (maximum likelihood with site-specific substitution rates) from the 5′ part of the genome, spanning bases 651 to 1600 of the alignment. (C) Representative tree (maximum likelihood with site-specific substitution rates) from the 3′ part of the genome, spanning bases 3001 to 4001 of the alignment. The sequences used are all available at http://hiv-web.lanl.gov by using either the common names or accession numbers (see Materials and Methods). The tree is unrooted. The scale shows the number of substitutions per site.
In order to examine individual regions of the entire genome, phylogenetic trees were constructed by using the maximum-likelihood method (Fig. 3B and C). The third-codon positions of the alignment were removed, and the alignment was gap stripped. Since phylogenetic trees spanning recombinant sequences are prohibited, the 4,411-bp remaining nucleotide alignment was divided into seven regions that did not contain any obvious recombination breakpoints in any of the aligned sequences (6). Two representative trees, one from the 5′ part and one from the 3′ part of the genome, are shown in Fig. 3B and C, respectively. SIVdrl1FAO, SIVmnd5440, and SIVmndM14 clustered together in all the phylogenetic trees built, and this relationship was supported in 100% of bootstrap replicates. As indicated by the short branch lengths, SIVmnd5440 and SIVmndM14 were extremely closely related and SIVdrl1FAO was more distantly related. In the 5′ tree, SIVmnd5440, SIVmndM14, and SIVdrl1FAO formed a cluster with SIVrcm. In the 3′ tree, however, SIVmnd5440, SIVmndM14, and SIVdrl1FAO grouped together with SIVmndGB1 and therefore clustered with the SIVlhoest lineage. This discordant topology was again indicative of recombination between an ancestor of SIVrcm and of SIVmnd GB1 (type 1). Phylogenetic trees including third-codon positions in the alignment showed the same topology as trees without third-codon positions.
Finally, in order to accurately delineate the recombination point where the relative relationship of SIVdrl1FAO or SIVmnd5440 to SIVrcm and SIVmndGB1 changed, we performed informative-site analysis and permutation tests (Fig. (Fig.4)4) (42). An informative site is one at which, in a four-sequence alignment, two sequences share one residue and the two others share another residue. The four-sequence alignment consisted of the potentially recombinant sequence (SIVdrl1FAO or SIVmnd5440, respectively), the two parental representative sequences (SIVrcm and SIVmndGB1), and an outgroup (SIVsyk). We considered only informative sites of first- and second-codon positions, since the majority of changes in the third positions are usually saturated in divergent sequences and do not contribute any reliable phylogenetic information. A total of 367 informative sites in SIVdrl1FAO and 358 sites in SIVmnd5440 were considered, and statistically significant breakpoints at the beginning of the envelope were observed for both SIVdrl1FAO (between nucleotides [nt] 2944 and 2945 of the alignment) and SIVmnd5440 (between nt 2939 and 2945). These data suggest with high probability that SIVdrl1FAO and SIVmnd5440 are descendents of the same recombinant.

Informative-site analysis. The recombination breakpoint in SIVdrl1FAO and SIVmnd5440 at which the closest relationship changed from SIVrcm to SIVmndGB1 was determined by informative-site analysis of first- and second-codon positions of a nucleotide alignment of SIVdrl1FAO or SIVmnd5440, SIVrcm, SIVmndGB1, and SIVsyk (outgroup) by surveying the informative sites supporting either of the two rooted phylogenies and maximizing a chi-square value to locate the optimal breakpoint location (33, 42). Statistical significance was assessed by performing 10,000 permutations and for each maximizing a chi-square value in order to again locate the optimal breakpoint. The P value reflects the proportion of permutated informative sites with chi-square values greater than the observed value. Total of 367 and 358 informative sites in SIVdrl1FAO and SIVmnd5440, respectively, were considered. A highly significant breakpoint was found between nt 2944 and 2945 for SIVdrl1FAO and between nt 2939 and 2945 for SIVmnd5440. Both breakpoints were located at the beginning of the envelope.
Phylogenetic relationship and origin of the vpx gene.
There are now several highly divergent SIV and HIV-2 strains that contain a vpx gene in addition to vpr. Previous studies have shown that the vpx and vpr genes of HIV-2, SIVsm, and SIVmac are phylogenetically related (46, 52). Recently, we described a novel PLV, SIVrcm, with a highly divergent vpx gene; it was only 42% identical on the amino acid level to SIVsm Vpx. As shown in Table Table2,2, SIVdrl1FAO Vpx was 70% related to SIVmnd5440 and SIVmndM14 Vpx and 57% related to SIVrcm Vpx. Figure Figure55 depicts an alignment of the major representatives of Vpx protein sequences, with the consensus sequence shown on top. Although the majority of the amino acids differ from each other, there was a highly conserved tyrosine motif in the central part of Vpx (Y-x-x-Y-R-Y-L-x-L). In addition, as already reported previously (32), most of the Vpx sequences, except that of SIVrcm, have a proline stretch at the C terminus.

Alignment of Vpx proteins from SIVmac251, HIV2/Ben, SIVsmPBj6.6, SIVrcmNG411, SIVmndM14, SIVmnd5440, and SIVdrl1FAO. The consensus is shown at the top. Dots indicate amino acid identity, and dashes indicate gaps introduced to optimize the alignment. Twenty-six conserved amino acids in the consensus sequence are in boldface and underlined.
In order to determine the phylogenetic relationships, we included all of the vpx sequences in a maximum-likelihood tree with published representatives of vpr sequences to investigate the origins of the vpx genes (Fig. (Fig.6).6). Despite being highly divergent, the vpx sequences formed a monophyletic cluster relative to vpr sequences. This supports a common origin of vpx genes found in the different vpx-bearing SIV strains and HIV-2, rather than the alternative that multiple vpr genes ultimately gave rise to the diverse forms of vpx. The SIVdrl1FAO/SIVmnd5440 vpx was more closely related to SIVrcm vpx than to SIVsm/HIV-2 vpx. As previously reported (46), the vpx gene cluster was phylogenetically most closely related to SIVagm vpr and did not closely group with SIVsm/HIV-2 vpr or with SIVrcm, SIVmnd, or SIVdrl vpr.

Phylogenetic analysis of vpx and vpr genes. A maximum-likelihood tree (Hasegawa-Kishino-Yano 1985 nucleotide substitution model [20a], codon position site-specific rates, tree bisection-reconnection branch swapping) was built from a vpx and vpr codon-based sequence alignment of 179 nucleotides (after excluding gaps and ambiguous nucleotides). The sequences used are all available from http://hiv-web.lanl.gov. Bootstrap support (100 replicates) was calculated by using the PHYLIP SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE programs. The tree is unrooted. The different lineages are annotated in boldface.
SIVdrl and SIVmnd5440 replicate in human PBMC and human T-cell lines but not in chimpanzee PBMC.
The replication potentials of SIVdrl1FAO and SIVmnd5440 were tested in a panel of human T-cell lines and human or chimpanzee PBMC. The human CD4+ cell lines included CEM-SS, CEMx174, C8166, H9, Hut-78, MT-2, MT-4, Molt4clone8, PM1, SupT1, and U937. The in vitro host ranges of SIVdrl1FAO and SIVmnd5440 are shown in Table Table4.4. Human PBMC supported replication of both SIVdrl1FAO and SIVmnd5440 with similar replication kinetics (Fig. (Fig.7A;7A; Table Table4).4). However, SIVdrl1FAO and SIVmnd5440, like SIVrcm, did not replicate in chimpanzee PBMC (Fig. (Fig.7B).7B). SIVmndGB1 served as positive control, since it has been shown previously to replicate to high titers in chimpanzee PBMC (6). SIVdrl1FAO also replicated in the CEMx174, CEM-SS, MT-4, Molt4clone8, and SupT1 cell lines whereas SIVmnd5440 replicated in CEMx174, Molt4clone8, and SupT1 cells (Table (Table4).4). No cytopathic effect was observed following infection of the susceptible cell lines or PBMC.

Comparative replication kinetics of SIVdrl1FAO and SIVmnd5440 in human and chimpanzee PBMC. PHA-stimulated human PBMC (A) and chimpanzee PBMC (B) were infected with equivalent amounts of virus (measured by RT as described in Material and Methods). Cell supernatants were collected at 3-day intervals and tested for RT activity. SIVrcmNG411 and SIVmndGB1 were used as positive controls for the replication in human and chimpanzee PBMC, respectively.
TABLE 4.
Host ranges of SIVdrl1FAO and SIVmnd5440 in different cell lines
| PBMC or cell line | RT activity of:
| |
|---|---|---|
| SIVdrl1FAO | SIVmnd5440 | |
| CEMx174 | + | + |
| CEMss | + | − |
| C8166 | − | − |
| H9 | − | − |
| Hut78 | − | − |
| MT-2 | − | − |
| MT-4 | + | − |
| Molt4 clone8 | + | + |
| PM1 | − | − |
| SupT1 | + | + |
| U937 | − | − |
| PBMC | ||
    Pig-tailed macaque Pig-tailed macaque | + | + |
    Chimpanzee Chimpanzee | − | − |
    Human Human | + | + |
The coreceptor usage of SIVdrl1FAO differs from those of SIVrcm and SIVmndGB1.
The coreceptor usage of SIVdrl1FAO was determined by using the GHOST cell assay. The GHOST cell lines constitute of a HIV-1-HIV-2-SIV indicator cell panel whose individual lines express a specific viral coreceptor molecule in conjunction with human CD4 (34). The coreceptor molecules included CCR1, CCR2B, CCR3, CCR4, CCR5, CCR8, CXCR4, GPR15 (BOB), and CXCR6 (STRL33, Bonzo). The production of SIV Gag p27 antigen in the supernatants collected at different time points post infection was measured. Figure Figure88 illustrates the time course of production of p27 antigen in the panel of GHOST cell lines. SIVdrl1FAO used CCR5, CXCR6 (STRL33, Bonzo), and GPR15 (BOB) as coreceptors for viral entry. This contrasted to the coreceptor usage of CCR2B and CXCR6 (STRL33, Bonzo) of SIVrcm and CXCR4 of SIVmndGB1 (6, 8, 43).

Evaluation of coreceptor usage of SIVdrl1FAO in the GHOST cell assay. Human osteosarcoma cells transfected with HIV-2 long terminal repeat-green fluorescent protein, CD4, and different HIV coreceptors were infected with SIV or HIV in the presence of Polybrene as indicated. The course of infection of GHOST cell lines was monitored by SIV Gag p27 antigen measurements in culture supernatants with the RETRO-TEK SIV-1 p27 antigen enzyme-linked immunosorbent assay (ZeptoMetrix Corp.) according to the manufacturer's instructions.
DISCUSSION
Recent studies (48, 50) have demonstrated that there are two different forms of SIV circulating in wild mandrills in Africa: SIVmnd type 1, represented by SIVmndGB1, and SIVmnd type 2, represented by SIVmndM14. SIVmnd type 1 clustered with SIVlhoest and SIVsun, whereas SIVmnd type 2 was related to SIVrcm in gag and pol and to SIVlhoest/sun in env and nef and carried a vpx gene in addition to vpr. In the present study, we showed that SIV from drill was most closely related to the recently described SIVmnd type 2 and also carried a vpx gene. In addition, we found a novel SIV from mandrill that was more closely related to SIVmndM14 than SIVdrl1FAO and therefore represented another SIVmnd type 2 isolate. Partial sequencing of a small portion of pol of two other mandrill isolates indicated that these isolates were also representatives of SIVmnd type 2.
Earlier phylogenetic analyses of SIVmndM14 were suggestive of recombination between SIVrcm and SIVmnd type 2, with the breakpoint estimated to be located in the vpr-tat region (48). Since our full-length sequence of SIVdrl1FAO was related to SIVmndM14, we wished to determine whether (i) SIVdrl was also recombinant between SIVrcm and SIVmnd type 1 and (ii) SIVdrl and SIVmnd type 2 were generated by the same recombination event. By applying informative-site analysis, a highly significant breakpoint was observed at the same location of the 5′part of the envelope in SIVdrl1FAO and SIVmnd5440 (Fig. (Fig.4).4). These results suggested that SIVdrl1FAO and SIVmnd5440 are descendents of the same recombinant. Either drills or mandrills may have been the original host for this type of virus, with the virus subsequently crossing the species barrier at some time in the past. Alternatively, the recombinant form could have been present in an ancestor of drill and mandrill. This scenario is highly unlikely, since SIVmnd type 2 is present only in mandrills in their northern habitat (south of the Sanaga river in Cameroon and north of the Ogooue river in Gabon) and not in mandrills in their southern habitat (south of the Ogooue river in Gabon). Interestingly, the habitat of SIVmnd type 2-infected mandrills is adjacent to but not overlapping with the drill habitat. An isolated population of drills also lives offshore on Bioko island (for drill and mandrill geographic distributions, see Fig. Fig.11 in reference 48). The relationship between SIVdrl1FAO from a captive drill and previously reported SIVdrl sequences from wild-caught drills (11, 48) indicated that SIVdrl occurs naturally in drill populations in the wild. Since the habitats of drill and mandrill are not sympatric, it is not clear how the cross-species transmission between mandrills and drills occurred. However, the present distribution of primates in Africa is likely to differ from that at the time when the recombination event occurred. The presence of recombinant forms in different primate species indicates not only that cross-species transmission occurs in wild populations but also that these events are not necessarily dead ends, since the transmitted forms can spread within the new host population. In addition, the unexpected finding of a SIVdrl like sequence (SIVmnd2-99CM32) in a mandrill supports the possibility of viral transmission between both species.
Previously we demonstrated that SIV from red-capped mangabeys carried a highly divergent vpx gene, which was only 42% related to SIVsm vpx at the amino acid level (6). SIVdrl1FAO and SIVmnd5440 were genetically related to SIVrcm and also carried a vpx gene. Therefore, we were interested in investigating the phylogenetic relationships of the vpx and vpr genes. An alignment of all available Vpx protein sequences revealed a highly conserved tyrosine motif (Y-x-x-Y-R-Y-L-x-L), which is shared with the human acetyl coenzyme A carboxylase 1 (Swiss-Prot accession no. Q13085). As recently discussed (32), most Vpx sequences, except that of SIVrcm, have a proline stretch at the C terminus. Proline-rich sequences have been shown to provide binding sites for SH3 domains. However, since SIVrcm is lacking the proline stretch, it seems to be dispensable for Vpx function.
Phylogenetic analysis of vpx and another accessory gene, vpr, showed that vpx from SIVrcm/drl/mnd-2 and vpx from SIVsm/HIV-2 were phylogenetically related despite being highly divergent. The expanded tree with numerous vpx and vpr sequences also confirmed the earlier observation of Sharp et al. (46) that the vpx gene is phylogenetically related to SIVagm vpr and not to SIVsm or SIVrcm vpr. Therefore, the presence of the extra gene most probably originated by nonhomologous recombination rather than by gene duplication as proposed by Tristem et al. (52, 53).
In terms of biological properties, SIVdrl1FAO and SIVmnd5440 replicated efficiently in human PBMC, which could allow the virus to establish persistent infections in humans. Indeed, epidemiological surveys revealed a case in Cameroon of a human infected by a virus serologically related to SIVmnd. This observation suggested that mandrills may represent a viral reservoir for humans, similar to sooty mangabeys in western Africa and chimpanzees in central Africa (48). Like SIVrcm, SIVdrl1FAO and SIVmnd5440 were not able to infect chimpanzee PBMC in our studies (6). Since SIVmnd type 1 (GB1) was able to replicate in chimpanzee PBMC, SIVdrl1FAO and SIVmnd5440 were more similar to SIVrcm in terms of their host range than to SIVmnd type 1, underlining the importance of the gag and pol genes in establishing productive infections (47).
In summary, we isolated SIV from one drill and three mandrills and sequenced the full-length genomes of SIVdrl1FAO and SIVmnd5440. Both isolates were genetically most closely related to SIVmndM14, the prototype of SIVmnd type 2. Phylogenetic and informative-site analyses showed that both SIVdrl and SIVmnd5440 were recombinant between ancestors of SIVrcm and SIVmnd type 1, with the recombination point located at the beginning of envelope. These data suggested with high probability that SIVdrl and SIVmnd type 2 were generated by the same recombination event. Finally, evaluation of the biological properties revealed that SIVdrl and SIVmnd5440 were able to replicate in human but not chimpanzee PBMC, indicating a potential risk of transmission upon contact between humans and infected primate specimens.
.
Acknowledgments
The GHOST cell panel was obtained from the NIH AIDS Research and Reference Reagent Program (provided by Vineet N. KewalRamani and Dan R. Littman). We thank Marisa St. Claire and Randy Elkins for providing blood samples from chimpanzees, Sonya Whitted for technical assistance, and the veterinary staffs of the zoos for providing drill and mandrill blood samples.
D.L.R. is supported by a Wellcome Trust Biodiversity Fellowship.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.77.8.4867-4880.2003
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc152139?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.77.8.4867-4880.2003
Article citations
HIV-2/SIV Vpx antagonises NF-κB activation by targeting p65.
Retrovirology, 19(1):2, 24 Jan 2022
Cited by: 4 articles | PMID: 35073912 | PMCID: PMC8785589
HUSH, a Link Between Intrinsic Immunity and HIV Latency.
Front Microbiol, 10:224, 12 Feb 2019
Cited by: 15 articles | PMID: 30809215 | PMCID: PMC6379475
Review Free full text in Europe PMC
The Expression of Functional Vpx during Pathogenic SIVmac Infections of Rhesus Macaques Suppresses SAMHD1 in CD4+ Memory T Cells.
PLoS Pathog, 11(5):e1004928, 21 May 2015
Cited by: 16 articles | PMID: 25996507 | PMCID: PMC4440783
Genetic characterization of near full length SIVdrl genomes from four captive drills (Mandrillus leucophaeus).
AIDS Res Hum Retroviruses, 31(3):353-357, 21 Jan 2015
Cited by: 0 articles | PMID: 25523403
Understanding restriction factors and intrinsic immunity: insights and lessons from the primate lentiviruses.
Future Virol, 9(5):483-497, 01 Jan 2014
Cited by: 5 articles | PMID: 26543491 | PMCID: PMC4630824
Go to all (34) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Genes & Proteins
- (1 citation) UniProt - Q13085
Nucleotide Sequences (Showing 14 of 14)
- (2 citations) ENA - AF301156
- (2 citations) ENA - M29975
- (2 citations) ENA - AF349680
- (2 citations) ENA - M27470
- (2 citations) ENA - AF075269
- (2 citations) ENA - L40990
- (2 citations) ENA - AY159322
- (2 citations) ENA - AJ310481
- (2 citations) ENA - AY159321
- (2 citations) ENA - L09212
- (2 citations) ENA - L06042
- (2 citations) ENA - U04005
- (2 citations) ENA - X52154
- (2 citations) ENA - AF328295
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Wild Mandrillus sphinx are carriers of two types of lentivirus.
J Virol, 75(15):7086-7096, 01 Aug 2001
Cited by: 87 articles | PMID: 11435589 | PMCID: PMC114437
Natural infection of wild-born mandrills (Mandrillus sphinx) with two different types of simian immunodeficiency virus.
AIDS Res Hum Retroviruses, 17(12):1143-1154, 01 Aug 2001
Cited by: 30 articles | PMID: 11522184
Phylogenetic analysis of SIV derived from mandrill and drill.
Front Biosci, 9:513-520, 01 Jan 2004
Cited by: 6 articles | PMID: 14766387
Review
SIVdrl detection in captive mandrills: are mandrills infected with a third strain of simian immunodeficiency virus?
Retrovirology, 1:36, 01 Nov 2004
Cited by: 1 article | PMID: 15516270 | PMCID: PMC529309
The history of SIVS and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa.
Front Biosci, 9:225-254, 01 Jan 2004
Cited by: 90 articles | PMID: 14766362
Review
Funding
Funders who supported this work.


