Abstract
Free full text

Evidence of Recombination between Divergent Hepatitis E Viruses
Abstract
Phylogenetic and recombination analysis was performed on 32 complete hepatitis E virus (HEV) genomes from infected humans and pigs. For the first time, evidence for recombination between divergent HEV strains was obtained, with at least two strains being found to have discordant phylogenetic relationships consistent with the occurrence of intragenotype recombination. This finding confirms that humans can be dually infected with divergent HEV strains and has implications for the emergence and evolution of new HEV epidemics.
Hepatitis E virus (HEV) is a causative agent of enterically transmitted hepatitis disease and, apart from infections of pregnant women, is generally associated with low mortality (11, 17). Outbreaks of HEV have been described to occur in many regions of the world, including Asia, Africa, and Mexico (1, 33, 53), while sporadic cases have been documented in the United States, Africa (9), and Europe (27, 37, 38, 60). HEV is presumed to be a zoonotic disease, as surveys of both wild and domestic animals have found evidence for human-related HEVs in rats (12, 22), pigs (2, 7, 21, 29, 32, 56), deer (45), and wild boar (41). Direct animal-to-human transmission of HEV has been demonstrated, for example, from deer to human as a result of eating uncooked meat (45). Interestingly, HEV characterized from swine in countries where HEV is not endemic is usually closely related to cases of sporadic human HEVs from the same country, for example, in the United States and Japan (29, 30, 42, 57), while in India, where HEV is endemic, human and swine HEV strains are not closely related (2). This suggests that in certain regions HEV is more established in the human population and that in other regions HEV is emerging as a result of direct contact with animal reservoirs for the virus. Divergent HEVs with no human counterparts have also been found in chickens (avian HEV). Avian HEV shares only 50 to 60% nucleotide sequence identity with HEV from humans and swine (15, 16).
HEV is a positive single-stranded RNA virus that is approximately 7,200 nucleotides (nt) in length. HEV's genome is capped and polyadenylated and has three open reading frames (ORF) (Fig. (Fig.1).1). As the organization of HEV's genome resembles that of Caliciviridae, at one time it was considered a member of this family (11). Sequence analysis has suggested that HEV should be reclassified, and the International Committee on Taxonomy of Viruses has created a new family, Hepeviridae, with HEV as the prototypic member of the genus Hepevirus (10). HEV has been classified further into four major genotypes (I to IV) (39). Genotype I includes Asian strains from India, Burma, Nepal, China, and Pakistan (3, 4, 13, 31, 40, 44, 49, 55) and African strains from Chad, Algeria, Tunisia, Morocco, Egypt, and Namibia (6, 18, 26, 48, 54). Genotype II includes U.S., Japanese, and European strains (38, 43, 60). Genotype III includes Mexican and African (Nigerian) strains (5, 20), and genotype IV includes Chinese (Shanghai) and Japanese strains (43). Note that alternative proposals for the classification of HEV strains have also been made (see the review by Schlauder and Mushahwar [39] for a detailed discussion).

Genomic organization of HEV. The scale at the bottom of the figure corresponds to nucleotides (in thousands). ORF1 encodes a nonstructural polyprotein which provides guanylyl-methyltransferase and RNA-dependent RNA polymerase activities and possibly other functions (23, 25). The ORF2 and ORF3 proteins are believed to be encoded by individual subgenomic RNAs generated during replication (44, 58). ORF2 encodes the viral capsid protein; ORF3, which contains one potential phosphorylation site, encodes a very small protein of about 123 amino acids (59). ORF3 proteins partition with the cytoskeleton, and it has been suggested that an interaction between ORF2 and ORF3 is associated with the assembly of virions (47, 50). ORF3 is also believed to interact with proteins involved in the host cell signaling pathway (24).
Analysis of the complete genome of a new African strain from Chad resulted in the proposal of a subgenotype of genotype I (or possibly a new HEV genotype) for that strain and an African strain from Morocco (52). Interestingly, the study of the Moroccan strain revealed the possibility of recombination between some of the genotype I strains. The aim of this study was to analyze the 32 available complete HEV genome sequences in order to systematically investigate the presence of recombination among HEV strains. A search for recombination events between the strains, within or between genotypes, was done using split decomposition, diversity plotting, an informative-site test, and statistical comparisons of maximum likelihood phylogenetic trees, as previously described (19, 35, 36).
The following 32 sequences were retrieved from GenBank: M73218-Burma, 7,194 nt; D10330-Myanmar, 7,194 nt; M74506-Mexico, 7,170 nt; M80581-Sar55/Pakistan, 7,138 nt; M94177-China B/Xinjiang, 7,194 nt; L25595-China C/Xinjiang, 7,193 nt; D11093-China D/Xinjiang, 7,194 nt; X99441-India/Madras, 7,194 nt; AF051830-Nepal/TK15-92, 7,193 nt; AF185822-Abb/2B Pakistan, 7,143 nt; AF060668-HEV US-1, 7,186 nt; AF060669-HEV US-2, 7,251 nt; AJ272108-Genotype 4/China Shanghai, 7,232 nt; AB074917-JKK Sap/Japan, 7,223 nt; AB074915-JAK Sai/Japan, 7,224 nt; AB074920-JMY-Haw/ Japan, 7,228 nt; AB074918-JKN Sap/Japan, 7,244 nt; AY230202-Morocco, 7,143 nt; AY204877-Chad T3, 7,154 nt; AF076239-India Hyderabad, 7,129 nt; X98292-fulminant India, 7,193 nt; AF082843-USA Swine, 7,207 nt; AB073912-swJ570/Japan, 7,225 nt; AB080575-HE-J14/Japan, 7,171 nt; AB089824-HE-JA10/Japan, 7,244 nt; AB097812-HE-JA1/Japan, 7,240 nt; AP003430-JRA1/Japan, 7,230 nt; AY115488-swCAN-Arkell/Canada, 7,242 nt; AF459438-North India/Jameel, 7,191 nt; L08816-China/Xinjiang, 7,176 nt; AB097811-swJ13-1/Japan, 7,258 nt; and AB108537-CCC220 China/Changchun, 7,193 nt. Multiple sequence alignments were constructed using the CLUSTALW program, version 1.8 (available from http://www-igbmc.u-strasbg.fr/BioInfo). The alignment was adjusted to be in the correct reading frame by using the alignment editor Se-Al (version 2.0; available from http://evolve.zoo.ox.ac.uk). Ambiguous regions and all sites including a gap were removed from the alignment.
First, a representation of the phylogenetic history of the 32-genome sequence alignment was inferred using split decomposition, implemented with the program SplitsTree 2.4 (available from http://www-ab.informatik.uni-tuebingen.de/software), with the pairwise differences between sequences estimated by the Kimura 3-ST model. Split decomposition depicts parallel edges between sequences if there are conflicting phylogenetic signals in the data. When the network was depicted to scale, one split regarding the phylogenetic position of genotype I strain China D was indicated (Fig. (Fig.2A).2A). Note that it is necessary to view the network without branch lengths in order to visualize all of the possible splits in the data. This revealed that, in addition to China D, the genotype I strains Nepal TK15, Morocco, and Chad T3 and the genotype IV strain HE-JA1 exhibited conflicting phylogenetic signals (Fig. (Fig.2B2B).

Network generated by split decomposition using the whole set of 32 complete HEV sequences (A) with and (B) without branch lengths included. Putative recombinant strains are indicated. The HEV genotypes (I to IV) are labeled.
To further investigate these putative recombinants, diversity plotting was performed on each by using a window of 300 nucleotides in increments of 50 nucleotides (available from http://bioinf.man.ac.uk/~robertson/recombination). Only two strains, China D and Nepal TK15, showed clear evidence for relative shifts in pairwise diversity with other strains (Fig. (Fig.3A3A and and4A,4A, respectively). Strain HE-JA1 does show a shift in the relative closeness of different genotype IV strains, but due to the low levels of diversity, this result is less conclusive. The results for the Morocco and 83-Chad T3 strains were inconclusive (data not shown). This could be due to false-positive results from the split-decomposition analysis or the absence of strains in our data set that are sufficiently related to the actual strains involved in the putative recombination. Diversity plotting was also performed on the remaining 27 sequences, and no further evidence of shifts in relative diversity indicative of recombination were observed. This indicates that split decomposition alone is an effective method for identifying (and visualizing) recombination in a data set.
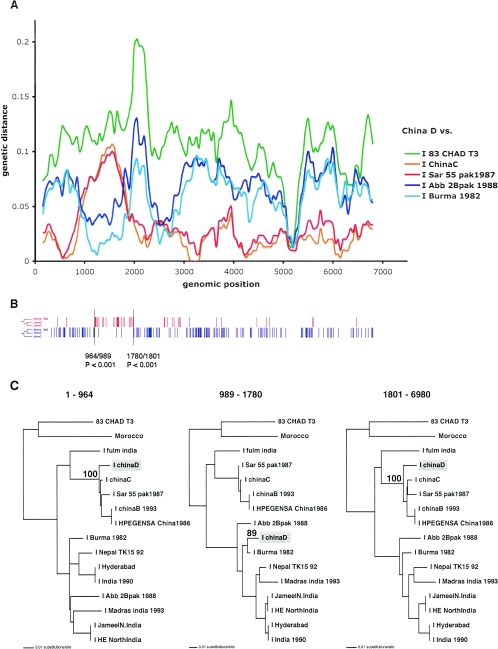
(A) Diversity plot comparing the strain China D to representative genotype I strains. The other comparisons are not shown for clarity. Pairwise genetic distances were calculated for a window of 300 nucleotides moved across the alignment in increments of 50 nucleotides. Genetic distance was plotted against the midpoint of each window for each comparison. (B) The distributions of the two types of informative sites in a four-sequence alignment that support the only two rooted tree topologies (shown to the left) are indicated in red and blue. The sequences included were the genotype I strains China D, Burma 1982, China C, and an outgroup, Chad T3. The positions of significant breakpoints, the numbers of the informative sites on either side of the breakpoint, and the P values are indicated. (C) Maximum likelihood phylogenies inferred for the three regions delimited by the informative-site analysis. Only bootstrap values pertaining to the clustering of China D are shown. The phylogenies are midpoint rooted. The scale corresponds to the number of nucleotide substitutions per site.

(A) Diversity plot comparing the strain Nepal TK15 to representative genotype I strains. (B) The distributions of the two types of informative sites in a four-sequence alignment that support the only two rooted tree topologies (shown to the left) are indicated in red and blue. The sequences included were the genotype I strains Nepal TK15, India 1999, Madras India 1993, and an outgroup, Chad T3. (C) Maximum likelihood phylogenies inferred for the three regions delimited by the informative-site analysis. Only bootstrap values pertaining to the clustering of the Nepal TK15 are shown. See the legend for Fig. Fig.33 for further details.
For three of the putative recombinants (China D, Nepal TK15, and HE-JA1), the HEV lineages that are relatively closely related to the respective “parental” strains involved in the recombination can be identified from the split-decomposition network (Fig. (Fig.2B).2B). This result was further confirmed by the diversity plotting (Fig. (Fig.3A,3A, ,4A,4A, and and5A).5A). As neither of these methods assessed the statistical support for the recombination, further analysis was performed. First, to more accurately locate the positions of the putative recombination breakpoints in China D, Nepal TK15, and HE-JA1 and to test their significance, a four-sequence informative-site test (4-SIS) was performed (available from http://bioinf.man.ac.uk/~robertson/recombination). The alignment included one of the recombinants, two genotype strains chosen from the split-decomposition analysis, and an outgroup sequence. See Fig. Fig.33 to to55 for details of the strains used for the analysis of China D, Nepal TK15, and HE-JA1, respectively. Two types of informative sites were distinguished, corresponding to the clustering of the putative recombinant with either of the reference strains chosen, and the distribution of these sites was surveyed for each (Fig. (Fig.3B,3B, ,4B,4B, and and5B).5B). The optimal breakpoint was located by maximizing a χ2 value (28, 35). Statistical significance was assessed by performing 1,000 permutations and for each permutation, maximizing the χ2 value. The P value reflects the proportion of permutated informative sites, with χ2 values being equal to or greater than the observed value. For China D, Nepal TK15, and HE-JA1 strains, a statistically significant breakpoint was located at positions 1780/1801, 4555/4684, and 2632/2974, respectively (Fig. (Fig.3B,3B, ,4B,4B, and and5B).5B). The 4-SIS test was repeated at either side of the first significant breakpoint, and for each strain, a second significant breakpoint was determined at positions 964/989, 581/940, and 4933/5058, respectively (Fig. (Fig.3B,3B, ,4B,4B, and and5B).5B). The 4-SIS test was repeated again on either side of these significant breakpoints. No further breakpoints were detected.

(A) Diversity plot comparing the strain HE-JA1 to representative genotype IV strains. (B) The distributions of the two types of informative sites in a four-sequence alignment that support the only two rooted tree topologies (shown to the left) are indicated in red and blue. The sequences included were the genotype IV strains HE-JA1, sw1311, HE-J14, and an outgroup, genotype 4. (C) Maximum likelihood phylogenies inferred for the three regions delimited by the informative-site analysis. Only bootstrap values pertaining to the clustering of HE-JA1 are shown. See the legend for Fig. Fig.33 for further details.
To further test the statistical support for recombination and to accurately infer the phylogenetic relationships of these apparently recombinant strains (China D, Nepal TK15, and HE-JA1), the breakpoints defined by the 4-SIS test were used to divide the alignments into three genomic regions, and phylogenetic trees were inferred by maximum likelihood analysis from these different regions by using PAUP* (Fig. (Fig.3C,3C, ,4C,4C, and and5C).5C). For each region, the HKY85 model of nucleotide substitution was used, incorporating an α shape parameter (using eight discrete categories of rate variation) for a gamma distribution of rate heterogeneity among sites, estimated from a neighbor-joining tree. The neighbor-joining tree topology was also used as the starting tree in a heuristic search using tree bisection and reconnection branch swapping for the maximum likelihood analysis. The reliability of the branching order was assessed by performing 1,000 bootstrap replicates with neighbor joining and the HKY85 model. Finally, the hypothesis that each recombinant clusters in different phylogenetic positions was tested for each phylogeny by constructing a tree with TreeEdit (available from http://evolve.zoo.ox.ac.uk), with the recombinant relocated to the alternative phylogenetic positions implicated in the recombination events. Statistical significance of these alternative phylogenetic hypotheses for each region was assessed with the one-tailed Kishino-Hasegawa test and the Shimodaira-Hasegawa test using PAUP* (data not shown). For each region of China D and Nepal TK15 that was tested, the maximum likelihood tree was the most significant (P < 0.05). These significantly discordant phylogenetic relationships confirm the existence of the two recombination breakpoints for each recombinant, as identified by the 4-SIS test. In addition, the bootstrap support values for the position of the recombinants China D and Nepal TK15 were high (Fig. (Fig.3C3C and and4C).4C). For HE-JA1, the bootstrap support value for the third region, positions 5058 to 6090, was low (65%), and the maximum likelihood tests were insignificant for all regions. Thus, we cannot definitively conclude that HE-JA1 is recombinant. However, it is intriguing that HE-JA1 was obtained from a 55-year-old Japanese man who lived in Sapporo City in Hokkaido, Japan, while swJ13-1 was obtained from a 4-month-old pig from a farm near Sapporo, Japan, indicating a geographical proximity for these infections of different host species (30).
In this study, we analyzed the available full-length genome sequences of HEV, carried out detailed recombination analysis, and definitively identified two recombinant HEV strains: China D and Nepal TK15 (Fig. (Fig.33 and and4,4, respectively). Note that China D was isolated in 1987 during the outbreak from 1986 to 1988 in the Xinjiang province in northwest China, while Nepal TK15 was isolated in 1992 in the Kathmandu region. The discordant phylogenetic relationships of these two strains demonstrate that recombination within HEV genotypes takes place, although no intergenotype recombination was detected. Recombination events must be the result of either coinfection or superinfection of the same infected host. Such dual infection of the same patient by HEV from two distinct genotypes has been documented (43). Similar recombination events have been described for other plus-strand RNA viruses, such as poliovirus, alphaviruses, and more recently, dengue virus (8, 14, 34, 46, 51). Presumably the occurrence of recombination and the detection of multiple breakpoints are due to divergent HEV genomes coming into proximity during replication and strand switching occurring.
Interestingly, there was some indication of recombination involving swine and human HEV strains from the split-decomposition and informative-site analyses. However, this result was not confirmed by all of the bootstrap support values or the maximum likelihood tests. This could be due to the conservative nature of these tests, coupled with the low diversity among these strains. However, we must conclude that, while the evidence is persuasive, it is by no means conclusive that HE-JA1 is recombinant. Nonetheless, these types of events involving recombination between viruses infecting different host species, even if tentative, need to be recorded, as they have serious implications for the future evolution of infectious agents, specifically for HEV's potential to emerge as a significant human disease and to become better established in the human population.
Acknowledgments
This work was supported by the French Army Health Service. J.F. received financial support from the University of Manchester OSS fund.
We are grateful to J. Viret, P. Binder, and D. Dormont for encouraging this research.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.79.14.9306-9314.2005
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1168788?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Genetic drift promotes and recombination hinders speciation on holey fitness landscapes.
PLoS Genet, 20(1):e1011126, 22 Jan 2024
Cited by: 0 articles | PMID: 38252672 | PMCID: PMC10833538
Viral hepatitis in pregnancy.
J Viral Hepat, 29(10):844-861, 07 Jul 2022
Cited by: 5 articles | PMID: 35748741 | PMCID: PMC9541692
Review Free full text in Europe PMC
Evolutionary Origins of Enteric Hepatitis Viruses.
Cold Spring Harb Perspect Med, 8(12):a031690, 03 Dec 2018
Cited by: 25 articles | PMID: 29610146 | PMCID: PMC6280709
Review Free full text in Europe PMC
Identification of a putative novel genotype 3/rabbit hepatitis E virus (HEV) recombinant.
PLoS One, 13(9):e0203618, 11 Sep 2018
Cited by: 7 articles | PMID: 30204796 | PMCID: PMC6133284
Hepatitis E virus infection of slaughtered healthy pigs in Brazil.
Zoonoses Public Health, 65(5):501-504, 13 Feb 2018
Cited by: 8 articles | PMID: 29441690
Go to all (29) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 32 of 32)
- (1 citation) ENA - AB097812
- (1 citation) ENA - AB108537
- (1 citation) ENA - AF185822
- (1 citation) ENA - AF459438
- (1 citation) ENA - AB097811
- (1 citation) ENA - AF082843
- (1 citation) ENA - M94177
- (1 citation) ENA - AB080575
- (1 citation) ENA - X99441
- (1 citation) ENA - AY230202
- (1 citation) ENA - X98292
- (1 citation) ENA - M73218
- (1 citation) ENA - M74506
- (1 citation) ENA - M80581
- (1 citation) ENA - AB074918
- (1 citation) ENA - AB074917
- (1 citation) ENA - AY204877
- (1 citation) ENA - AB074915
- (1 citation) ENA - D10330
- (1 citation) ENA - AJ272108
- (1 citation) ENA - AB073912
- (1 citation) ENA - AB074920
- (1 citation) ENA - AF051830
- (1 citation) ENA - L08816
- (1 citation) ENA - AF076239
- (1 citation) ENA - AY115488
- (1 citation) ENA - AB089824
- (1 citation) ENA - L25595
- (1 citation) ENA - AF060668
- (1 citation) ENA - AF060669
- (1 citation) ENA - AP003430
- (1 citation) ENA - D11093
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Partial nucleotide sequencing of hepatitis E viruses detected in sera of patients with hepatitis E from 14 cities in China.
Chin Med J (Engl), 115(7):1058-1063, 01 Jul 2002
Cited by: 11 articles | PMID: 12150743
Analysis of complete genome sequences of swine hepatitis E virus and possible risk factors for transmission of HEV to humans in Korea.
J Med Virol, 82(4):583-591, 01 Apr 2010
Cited by: 47 articles | PMID: 20166181
Ancient recombination events and the origins of hepatitis E virus.
BMC Evol Biol, 16(1):210, 12 Oct 2016
Cited by: 19 articles | PMID: 27733122 | PMCID: PMC5062859
Genetic variability and evolution of hepatitis E virus.
Virus Res, 127(2):216-228, 23 Mar 2007
Cited by: 208 articles | PMID: 17363102
Review
Hepatitis E virus.
Rev Med Virol, 13(3):145-154, 01 May 2003
Cited by: 299 articles | PMID: 12740830
Review

